RESEARCH & Technology development: PLATFORM
The aim of the functionalgenomics.pl since 2010 is to conduct stem cell research, within human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) to study human neurodevelopmental processes and disease under unique genetic patient-specific background.
Our patients’ specific and engineered stem cells serve as a platform for development of personalized gene therapies, compounds screening and drug repurposing. Cell fate reprogramming methods and differentiation of primary cells, including skin fibroblasts (derived by skin punch biopsies) and blood (peripheral blood mononuclear cells; PBMCs) are applied in the process.
The field of stem cell-based approaches has impressively progressed during past few years from modeling to treatment of the disease > see: Innovations in Stem Cell Biology Collection 2024. Below you can find figures depicting general modeling workflow and levels of the complexity applied in our stem cell based disease modeling platform:
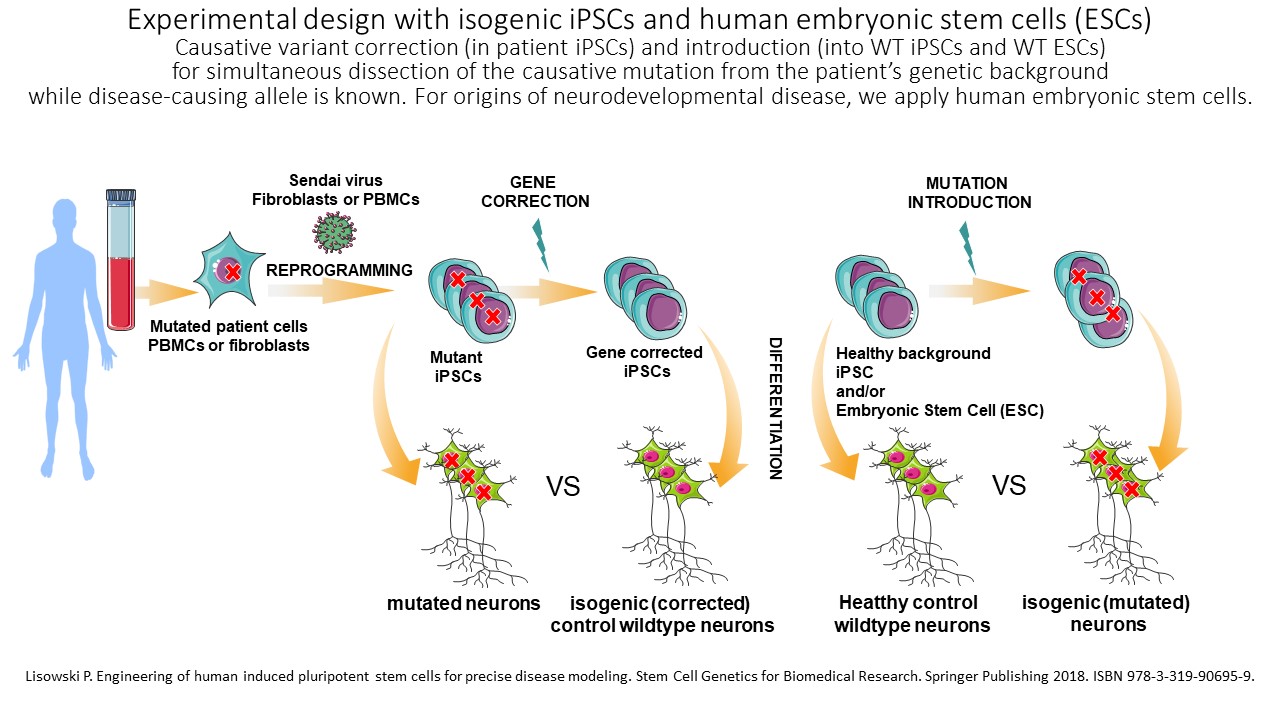
Generation of isogenic, patient and gene variant specific stem cell lines for precise disease modeling applied in our study:
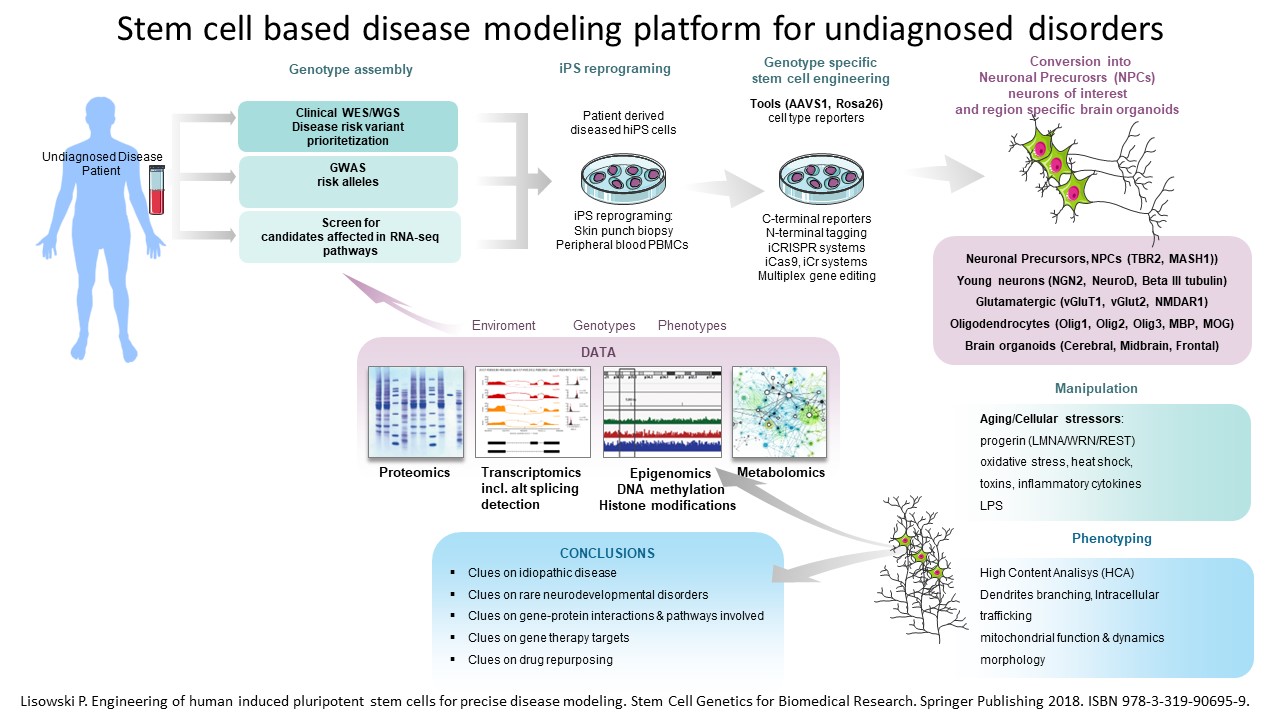
We developed stem cell based disease modeling technological platform to support clinical WES and WGS for diagnostics of Undiagnsosed Neurodevelopmental Disorders (UNDs)
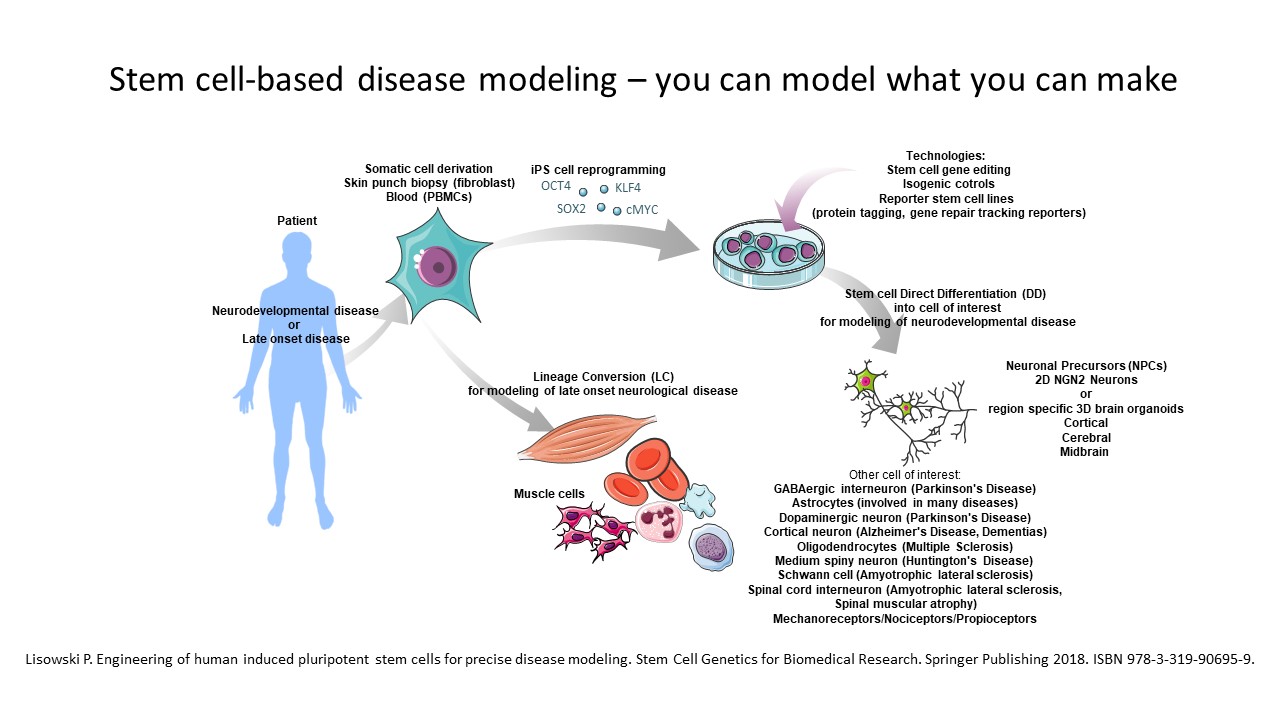
Examples of cell fate reprograming and differentiation for neurodevelopmental study or direct lineage conversion for late onset phenotypic study applied in our technological platform:
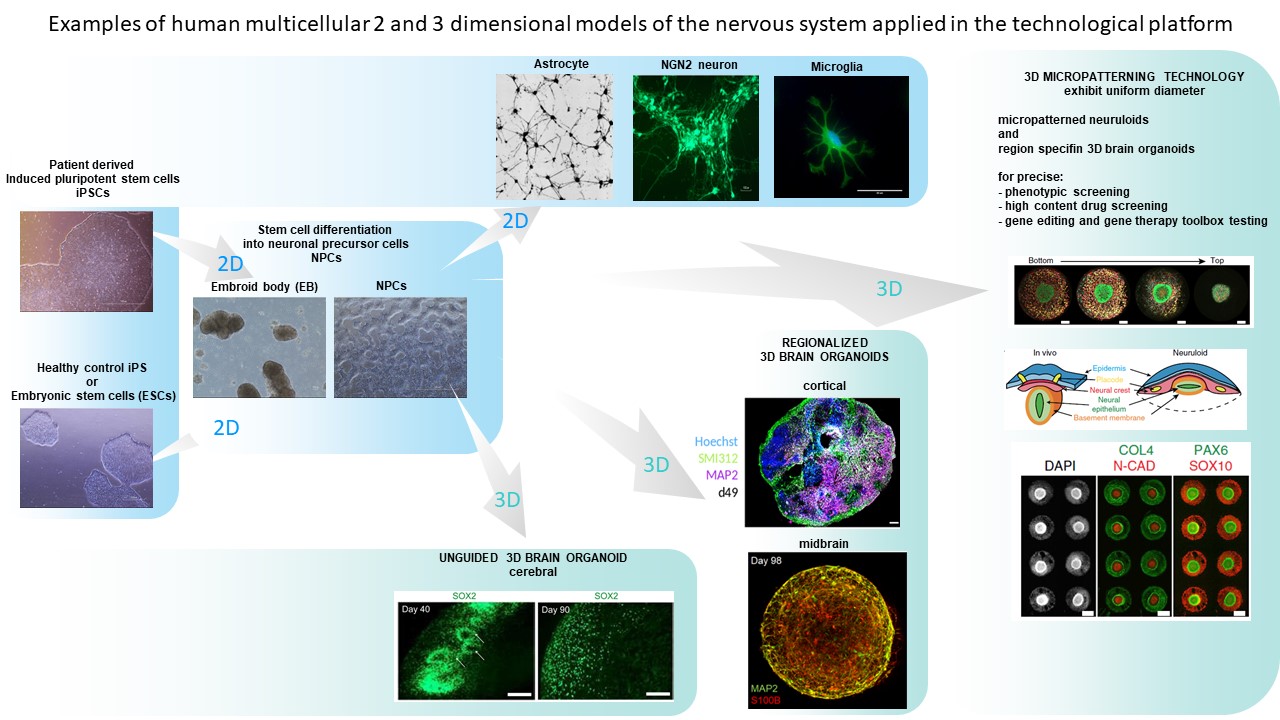
Following technological topics are covered to get molecular insight into neurodevelopmental, neurological and neuropsychiatric diseases:
In the field stem cell based disease modeling including service providing:
- Generation of patient specific induced pluripotent stem cells (hiPSCs) from blood and skin and their quality control (Q/C).
- Differentiation of patient stem cells into neurons of interest, brain organoids and micropatterned neuronal cells for high througput phenotypic, drug screening and gene therpy tools testing.
- Gene editing of stem cell for streamline production of isogenic cell lines for precise disease modeling (introductions and corrections of gene variants, generation of isogenic cell lines)
- Somatic cell genome editing (post mitotic neurons and astrocytes).
- Engineering of patient-specific stem cells towards generation of reporter cell lines (N-terminal biallelic tagging, C-terminal biallelic reporters)
- Extensive phenotyping (scRNAseq, spatial transcriptomics, AIR-alternative isoform regulation, mRNA isoforms analysis, shifts in splicing, Hight Content Screening, proteomics and metabolomics) along the neuronal lineage to unravel diseased phenotypes.
- Mapping of differentially expressed genes into the functional pathways and networks reflecting pathological events for identification of novel modules for high content drug screening.
- Conversion of patient specific or engineered stem cells into cells of interest for high throughput drug screening (drug repurposing).
In the field of genome editing technology development:
- Enhancing precise genome engineering by manipulation of DNA repair pathways and identification of the main factors shifting DSB repairs towards HDR pathway.
- Searching for alternatives of HDR for in vivo genome editing in non-dividing somatic cells to develop quality tools for effective and safe genome editing in human patients as somatic gene therapy.
- Optimization of somatic cell gene editing tolbox: adenine base editors, cytozine base editors (ABEs, CBEs) and prime editors (PEs).
- Development gene editing machinery delivery methods into somatic cell (rAAVs, VLPs, LNPs).
- Development of strategies and tools to monitor and/or reduce off target effects.
- Development of homology independent target replacement – repair of large mutations in somatic cell.
Unknown, rare and ultra-rare diseases diagnostics:
- Rare diseases are conditions that affect less than 1 in 2000 people in the population.
- There are >7000 identified rare diseases affecting millions people worldwide with genetic background and they manifest early on in life leading to severe health consequences.
- In >60% of the cases in which a rare genetic disease is suspected, whole exome (WES) and whole genome (WGS) sequencing fails to identify coding variants that are potentially pathogenic.
- Rare variants that affect gene regulation are expected to underlie pathogenesis in such cases.
Despite progress in technology while the genetic diagnosis cannot be reached we introduced the concept of combining the WGS data with the transcriptome and methylome study as a promising approach toward better understanding the extensive data generated by WGS and therefore help prioritize genetic alterations. RNA-Seq data can reveal diverse transcriptional changes, not only splice site mutations but also silent mutations in coding regions, deep intronic mutations, and/or structural changes causing splicing aberrations, impossible to interpret using the WGS data only. Comparison and integrated analysis of the RNA-Seq results with the methylome analysis and whole-genome sequence (WGS) can provide new evidence that uncovered genomic mutations trigger those transcriptional changes and underlie neurological disease under investigation. Therefore, by taking account of genomic mutations causing transcriptional aberrations, we can improve the sensitivity of deleterious mutation detection in the entire genome. Below figure depicts our stem cell-based DNA and RNA sequencing diagnostic workflow:
Above workflow aims to develope methods for exploration of trasncriptomic data to increase diagnostic yield for unknown and rare disease patients. It allowes for identification of genome regulatory regions responsible for aberrant expression of particular genes or group of genes in the most appropriate cells of particular patient. Such approach allows for prioritization of the genetic variants causing alterations in the transcript triggered by genetic aberrations of gene regulation.